…
New Look At Judas Gameplay Reveals More BioShock Inspiration
Ghost Story Games, the studio led by BioShock creator Ken Levine, revealed its first game, Judas, during The Game Awards 2022 in a cinematic trailer. Now, more than a year since that announcement, Ghost Story Games has revealed a new look at Judas gameplay. And perhaps unsurprisingly, it reminds us of BioShock (and it looks great).
Revealed in a new trailer that aired during today’s Sony State of Play, this look at Judas pulled back the curtains on what exactly is going on in this single-player, narrative-driven sci-fi game. Well, kinda. We got another look at the game’s interesting world, which shows Judas stuck on a space station with various characters (perhaps enemies?) trying to fix something that she might have broken. As for what that entails, it remains a mystery, but this new trailer sheds a little light on her mission.
Check it out for yourself in the new Judas gameplay trailer below:
[embedded content]
In a press release, Ghost Story Games says this trailer “offers a deeper look at the game’s setting aboard the Mayflower, a spacefaring city whose citizens are trained to tear each other apart for even the most minor infractions, and where machines control every aspect of business, art, and government.” As Judas, players are the drivers of every event in a mysterious story “with a new cast of characters to get to know – and to change – in a world where every decision you make affects how the story unfolds.”
Here’s the synopsis for Judas:
“You are the mysterious and troubled Judas. Your only hope for survival is to make or break alliances with your worst enemies. Will you work together to fix what you broke – or will you leave it to burn?”

Levine formed Ghost Story Games in 2014 after he closed BioShock Infinite studio, Irrational. He took a handful of Irrational developers with him to Ghost Story Games, where the team has been working for years on something involving “narrative legos.” Now, given how often things change in game development, it remains unclear if Judas is what the team has been working on for 10 years or not. In 2022, it was reported that the team’s first project had gone through a rocky development patch – you can read about that here.
Judas does not have a release date. It’s expected to launch on PlayStation 5, Xbox Series X/S, and PC, and will be published by Take-Two Interactive, which also published all three BioShock games.
For more about the game, head to Game Informer’s Judas tag here.
Are you excited about Judas? Let us know in the comments below!
The all-star AI-powered, cloud-delivered security platform you need – CyberTalk

EXECUTIVE SUMMARY:
Cyber criminals are weaponizing advanced tactics, including intricate social engineering campaigns, to carry out malicious activities.
Last year, social engineering attempts rose by 464%. Often a result of social engineering attempts, ransomware attacks have reportedly increased by 90%.
Amidst this unsettling attack landscape, it can be difficult and stressful to secure data and networks effectively.
Cyber security in 2024
“Leveraging AI to drive better security outcomes is top of mind for CISOs, as they address both the expanding threat landscape and the cyber security talent shortage,” says Frank Dickson, Group Vice President, Security & Trust, IDC.
“When selecting an AI-powered cyber security solution, CISOs are looking for a return on investment through increased productivity and better efficacy,” says Dickson.
Check Point Infinity Platform
To address these relentless challenges and the latest, contemporary cyber security considerations, Check Point has just revealed its unified and comprehensive security solution – The Check Point Infinity Platform.
This advanced platform marks a new era in AI-powered, cloud-delivered cyber security, as the platform is specifically designed to meet the modern challenges of an evolving threat landscape.
“Embracing the future of cyber security, we’re pioneering an AI-powered, cloud-delivered platform that embodies the convergence of intelligence and accessibility,” said Gil Shwed, CEO & Founder, Check Point Software Technologies at CPX 2024.
“This platform is not just a solution but a revolution, leveraging decades of R&D to offer real-time, sophisticated defense mechanisms. It represents a leap toward a more secure, agile, and interconnected digital landscape, where protection is not just reactive, but predictive and proactive.”
AI Infinity Copilot
Integrating seamlessly across the Check Point Infinity Platform, Check Point is also introducing Infinity AI Copilot, a tactical assistant that’s infused with the power of generative AI.
This tooling can automate tasks, reduce the time required for routine tasks, and provide proactive solutions to cyber security threats.
Key capabilities of Infinity AI Copilot
1. Enhance security administration efficiency. Infinity AI Copilot can reduce the administrative workload for cyber security tasks by up to 90%. It can assist with event analysis, implementation and troubleshooting.
2. Streamline security policy management and deployment. Infinity AI Copilot can effortlessly manage, modify and automatically deploy access rules and security controls that are tailored to individual policies.
3. Boost incident mitigation and response. Utilize this AI for threat hunting, analysis and resolution, along with enhancing the effectiveness of incident management.
4. Oversee the entire solution landscape. AI Copilot seamlessly oversees all products within the complete Check Point Infinity Platform, serving as a competent, comprehensive assistant.
5. Enables natural language processing simplicity. Interacting with Infinity AI Copilot will seem similar to a natural conversation with a human. The tool ‘understands’ and responds via chat in nearly any language, ensuring a smooth interaction and efficient task completion.
Infinity AI Copilot is currently available in preview mode, with a full launch expected in Q2. Future developments include proactive assistance and autonomous policy management features.
Get the full story here.
Looking for more info about the latest cyber security solutions? Join us for CPX 2024. Register now.
Professor Emeritus Igor Paul, an expert in product design and safety, dies at 87

Professor Emeritus Igor Paul ’60, SM ’61, PhD ’64, an influential professor of mechanical engineering, passed away on Dec. 17, 2023 at his home in St. Petersburg, Florida. He was 87.
Paul was a member of the MIT Department of Mechanical Engineering faculty from 1964 until his retirement in 2003, and helped to develop the department’s design and manufacturing curriculum, which continues to thrive today. His research interests included product and machine design, safety, and risk analysis; robotics; biomechanics; and dynamic systems modeling.
A leading expert in product design and safety, with a particular focus on sports devices like helmets, Igor served as an expert witness in many landmark product liability cases. He also contributed to the development of artificial joints and the development of inertial guidance systems for NASA and provided consulting services to a number of area hospitals and medical centers.
Paul was known for his good nature, quick wit, and pleasant disposition, and his deep passion for teaching. Among the courses he instructed through the years were 2.72 (Elements of Mechanical Design), 2.70 (now 2.007, Design and Manufacturing I), and 2.009 (Product Engineering Processes). He served for many years as the faculty advisor to the student chapter of the American Society of Mechanical Engineers.
He also co-authored more than 80 publications and won numerous awards in the areas of design, bio-engineering, and education, including the DeFlorez Award for Creativity in Design (MIT, 1960); the Ralph R. Teetor Distinguished Educator Award (SAE); Outstanding Orthopedic Research Award (Orthopedic Research Society); and the Carl Soderberg Distinguished Service Award (MIT, 2003).
Paul was born on Oct. 28, 1936, in Kharkov, Ukraine, and migrated across Europe during World War II, arriving in the United States eight years later on Christmas Day 1951. After gaining admission to MIT, he earned all three of his degrees in the Department of Mechanical Engineering.
He is survived by his wife, Natasha Paul (Gruzinov); his daughter, Tahisa Southwell of Las Vegas, Nevada; his son, Victor Paul of Zurich, Switzerland; and four grandchildren. He was preceded in death by his parents, Leo and Lily Paul; his sister, Nina Karouna; and his beloved daughter, Tanya Paul.
Outside of his professional achievements, Paul enjoyed tennis, golf, and traveling the globe. After his retirement in 2003, he and Natasha moved from Andover, Massachusetts, to New London, New Hampshire, then recently settled in St. Petersburg, Florida.
Paul leaves behind a legacy of scientific contributions, dedication to education, and love for his family.
MIT Press’s Direct to Open opens access to full list of 2024 monographs

Now in its third year of operation, the MIT Press’ Direct to Open (D2O) recently announced that it reached its full funding goal in 2024 and will open access to 79 new monographs and edited book collections this year.
Launched in 2021, D2O is an innovative sustainable framework for open-access monographs that shifts publishing from a solely market-based, purchase model where individuals and libraries buy single e-books, to a collaborative, library-supported open-access model.
“Reaching our overall funding goal — in full and on time — is a major milestone in developing a sustainable open-access publishing model,” says Amy Harris, senior manager, library relations and sales at the MIT Press. “We are extremely grateful for the support of our library and consortium partners that makes this possible.”
There are other models that offer fund-to-open opportunities on a title-by-title basis or that focus on opening access within specific disciplines. D2O is unique because it allows the press to open access to its entire slate of scholarly books at scale during each funding cycle. Thanks to D2O, all monograph authors have the opportunity for their work to be open access, and the press can offer equal support to traditionally underfunded disciplines in the social sciences and humanities.
At a time when the traditional market for scholarly books continues to decline, works funded through D2O are reaching larger audiences online than ever before — averaging 2,694 reads per title and bringing important scholarship to new audiences. D2O books have also been academically cited almost 1,100 times.
“D2O is meeting the needs of academics, readers, and libraries alike, and our usage and citation stats demonstrate that the academic community is embracing open-access scholarship across a wide range of fields and for many purposes — from the classroom to research projects to professional interest reading,” says Harris. “This further aligns the work of the MIT Press with the mission of MIT to advance knowledge in science, technology, the arts, and other areas of scholarship to best serve the nation and the world, and provides opportunities for expansion of the model in the forthcoming years.”
The MIT Press will now turn its attention to its fourth funding cycle and invites libraries and library consortia to participate. For details, please visit: mitpress.mit.edu/D2O.
New fellowship to help advance science journalism in Africa and the Middle East

The Knight Science Journalism Program at MIT has announced a new one-semester fellowship — the Fellowship for Advancing Science Journalism in Africa and the Middle East — that will start this year.
The fellowship, developed through a generous gift from the global publishing company Springer Nature, was created in honor of the influential Egyptian science journalist Mohammed Yahia, who died last year at the age of 41.
Yahia worked for Springer Nature for over 13 years, primarily as managing editor of the Nature Portfolio in the Middle East, where he built an award-winning team. He was widely admired for his work advancing the status of science journalism both in that region and throughout Africa. He was president of the World Federation of Science Journalists from 2017 to 2019, working also to help build a network of science journalists around the globe.
Springer Nature, the founding sponsor of the fellowship, is well-known for its standing as a publisher of some the most high-profile and respected research journals and magazines in the world. “Mohammed was known for his unwavering commitment to science and his talent for simplifying complex research,” says Stephen Pincock, vice president in Springer Nature’s Solutions Group. “With this fellowship we want to inspire more to follow in his footsteps, as trusted communicators of evidence-based research.”
The first Fellowship for Advancing Science Journalism in Africa and the Middle East will be hosted by the Knight Science Journalism Program this fall and will continue in subsequent fall semesters. Thanks to a generous grant from Springer Nature, the program will offer a $40,000 stipend for the fellowship period from Aug. 16 to Dec. 31. KSJ will also cover the fellow’s health insurance and a $5,000 housing stipend to help with relocation costs.
The Knight Science Journalism Program, established at MIT in 1983, is the world’s leading science journalism fellowship program. More than 400 leading science journalists from six continents have graduated from the full-year academic program, which offers a course of study at MIT, Harvard University, and other leading institutions in the Boston area, as well as specialized training workshops, seminars, and science-focused field trips for all attendees.
“The Knight Science Journalism Program is honored to partner with Springer Nature in honoring Mohammed Yahia and in creating this new fellowship to help support science journalism in this important part of the world,” says KSJ Director Deborah Blum. “We believe strongly in the global nature of both science and the importance of telling its story in the most helpful and insightful way. We believe this new fellowship is an excellent way to advance that mission.”
Fellows supported by this new program will join the regular KSJ class of journalists for the fall semester in a program of study at MIT and other Cambridge/Boston area universities and in the program’s seminars, training workshops, and field trips throughout the semester. They will also have access to such benefits as MIT’s program of subsidized public transportation and access to libraries, museums, and other Boston-area programs, as well as connections to a thriving community of science journalists.
The program will open an applications process for journalists from Africa and the Middle East on Feb. 1 and submissions will be accepted until March 1. All journalists from the region with at least three years of experience in covering science, health, and the environment are encouraged to apply. The selected fellow will be announced by the end of March.
For further questions about the fellowship or the application process, please write to info@ksj.mit.edu.
Blood cell family trees trace how production changes with aging
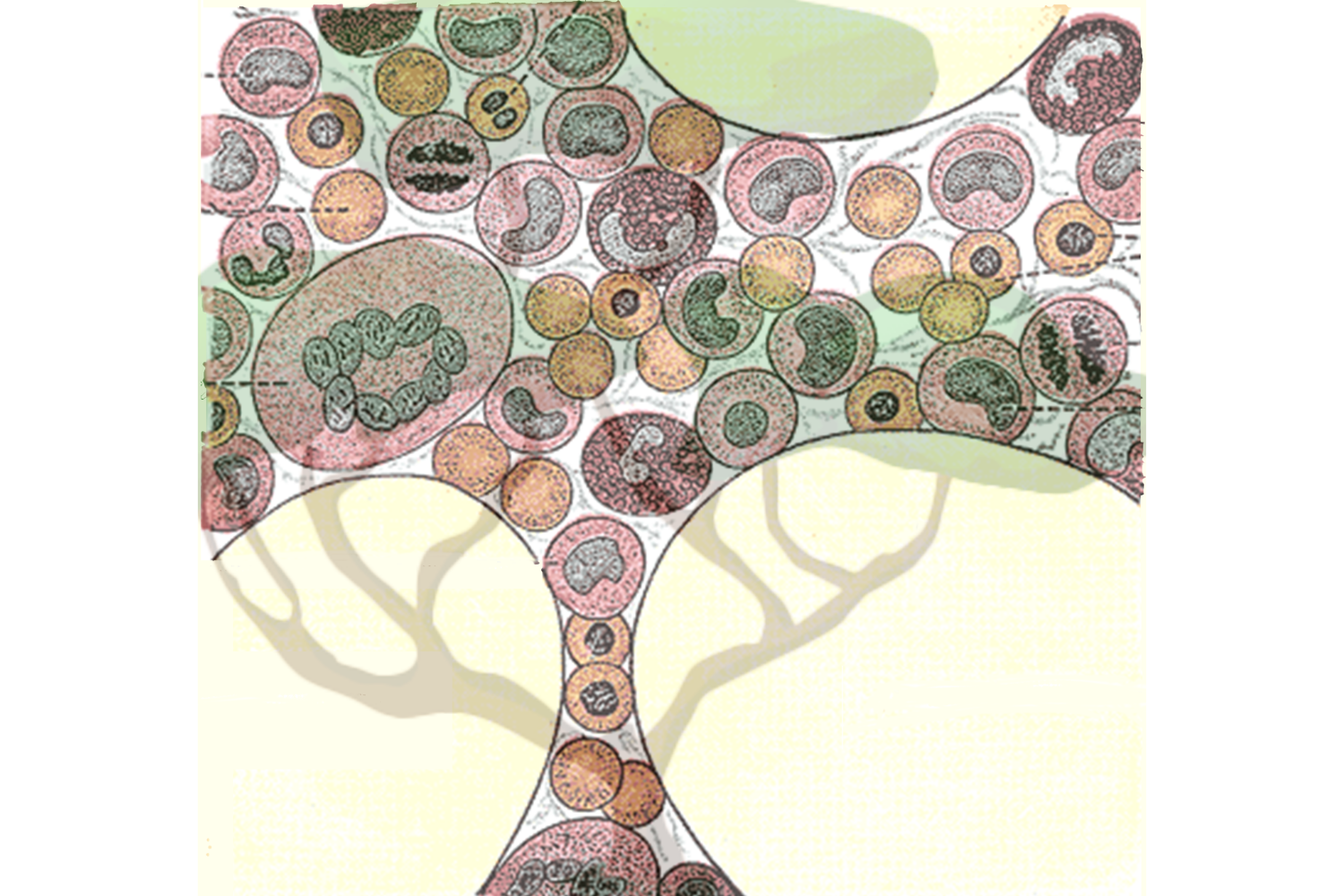
Blood cells make up the majority of cells in the human body. They perform critical functions and their dysfunction is implicated in many important human diseases, from anemias to blood cancers like leukemia. The many types of blood cells include red blood cells that carry oxygen, platelets that promote clotting, as well as the myriad types of immune cells that protect our bodies from threats such as viruses and bacteria.
What these diverse types of blood cells have in common is that they are all produced by hematopoietic stem cells (HSCs). HSCs must keep producing blood cells in large quantities throughout our entire lives in order to continually replenish our bodies’ supply. Researchers want to better understand HSCs and the dynamics of how they produce the many blood cell types, both in order to understand the fundamentals of human blood production and to understand how blood production changes during aging or in cases of disease.
Jonathan Weissman, an MIT professor of biology, member of the Whitehead Institute for Biomedical Research, and a Howard Hughes Medical Investigator; Vijay Sankaran, a Boston Children’s Hospital and Harvard Medical School associate professor who is also a Broad Institute of MIT and Harvard associate member and attending physician at the Dana Farber Cancer Institute; and Chen Weng, a postdoc in both of their labs, have developed a new method that provides a detailed look at the family trees of human blood cells and the characteristics of the individual cells, providing new insights into the differences between lineages of HSCs. The research, published in the journal Nature on Jan. 22, answers some long-standing questions about blood cell production and how it changes as we age. The work also demonstrates how this new technology can give researchers unprecedented access to any human cells’ histories and insight into how those histories have shaped their current states. This will render open to discovery many questions about our own biology that were previously unanswerable.
“We wanted to ask questions that the existing tools could not allow us to,” Weng says. “This is why we brought together Jonathan and Vijay’s different expertise to develop a new technology that allows us to ask those questions and more, so we can solve some of the important unknowns in blood production.”
How to trace the lineages of human cells
Weissman and others have previously developed methods to map the family trees of cells, a process called lineage tracing, but typically this has been done in animals or engineered cell lines. Weissman has used this approach to shed light on how cancers spread and on when and how they develop mutations that make them more aggressive and deadly. However, while these models can illuminate the general principles of processes such as blood production, they do not give researchers a full picture of what happens inside of a living human. They cannot capture the full diversity of human cells or the implications of that diversity on health and disease.
The only way to get a detailed picture of how blood cell lineages change through the generations and what the consequences of those changes are is to perform lineage tracing on cells from human samples. The challenge is that in the research models used in the previous lineage tracing studies, Weissman and colleagues edited the cells to add a trackable barcode, a string of DNA that changes a little with each cell division, so that researchers can map the changes to match cells to their closest relatives and reconstruct the family tree. Researchers cannot add a barcode to the cells in living humans, so they need to find a natural one: some string of DNA that already exists and changes frequently enough to allow this family tree reconstruction.
Looking for mutations across the whole genome is cost-prohibitive and destroys the material that researchers need to collect to learn about the cells’ states. A few years ago, Sankaran and colleagues realized that mitochondrial DNA could be a good candidate for the natural barcode. Mitochondria are in all of our cells, and they have their own genome, which is relatively small and prone to mutation. In that earlier research, Sankaran and colleagues identified mutations in mitochondrial DNA, but they could not find enough mutations to build a complete family tree: in each cell, they only detected an average of zero to one mutations.
Now, in work led by Weng, the researchers have improved their detection of mitochondrial DNA mutations 10-fold, meaning that in each cell they find around 10 mutations — enough to serve as an identifying barcode. They achieved this through improvements in how they detect mitochondrial DNA mutations experimentally and how they verify that those mutations are genuine computationally. Their new and improved lineage tracing method is called ReDeeM, an acronym drawing from single-cell “regulatory multi-omics with deep mitochondrial mutation profiling.” Using the method, they can recreate the family tree of thousands of blood cells from a human blood sample, as well as gather information about each individual cell’s state: its gene expression levels and differences in its epigenome, or the availability of regions of DNA to be expressed.
Combining cells’ family trees with each individual cell’s state is key for making sense of how cell lineages change over time and what the effects of those changes are. If a researcher pinpoints the place in the family tree where a blood cell lineage, for example, becomes biased toward producing a certain type of blood cell, they can then look at what changed in the cells’ state preceding that shift in order to figure out what genes and pathways drove that change in behavior. In other words, they can use the combination of data to understand not just that a change occurred, but what mechanisms contributed to that change.
“The goal is to relate the cell’s current state to its past history,” Weissman says. “Being able to do that in an unperturbed human sample lets us watch the dynamics of the blood production process and understand functional differences in hematopoietic stem cells in a way that has just not been possible before.”
Using this approach, the researchers made several interesting discoveries about blood production.
Blood cell lineage diversity shrinks with age
The researchers mapped the family trees of blood cells derived from each HSC. Each one of these lineages is called a clonal group. Researchers have had various hypotheses about how clonal groups work: Perhaps they are interchangeable, with each stem cell producing equivalent numbers and types of blood cells. Perhaps they are specialized, with one stem cell producing red blood cells, and another producing white blood cells. Perhaps they work in shifts, with some HSCs lying dormant while others produce blood cells. The researchers found that in healthy, young individuals, the answer is somewhere in the middle: Essentially every stem cell produced every type of blood cell, but certain lineages had biases toward producing one type of cell over another. The researchers took two samples from each test subject four months apart, and found that these differences between the lineages were stable over time.
Next, the researchers took blood samples from people of older age. They found that as humans age, some clonal groups begin to dominate and produce a significantly above-average percent of the total blood cells. When a clonal group outcompetes others like this, it is called expansion. Researchers knew that in certain diseases, a single clonal group containing a disease-related mutation could expand and become dominant. They didn’t know that clonal expansion was pervasive in aging even in seemingly healthy individuals, or that it was typical for multiple clonal groups to expand. This complicates the understanding of clonal expansion but sheds light on how blood production changes with age: The diversity of clonal groups decreases. The researchers are working on figuring out the mechanisms that enable certain clonal groups to expand over others. They are also interested in testing clonal groups for disease markers to understand which expansions are caused by or could contribute to disease.
ReDeeM enabled the researchers to make a variety of additional observations about blood production, many of which are consistent with previous research. This is what they hoped to see: the fact that the tool efficiently identified known patterns in blood production validates its efficacy. Now that the researchers know how well the method works, they can apply it to many different questions about the relationships between cells and what mechanisms drive changes in cell behavior. They are already using it to learn more about autoimmune disorders, blood cancers, and the origins of certain types of blood cells.
The researchers hope that others will use their method to ask questions about cell dynamics in many scenarios in health and disease. Sankaran, who is a practicing hematologist, also hopes that the method one day revolutionizes the patient data to which clinicians have access.
“In the not-too-distant future, you could look at a patient chart and see that this patient has an abnormally low number of HSCs, or an abnormally high number, and that would inform how you think about their disease risk,” Sankaran says. “ReDeeM provides a new lens through which to understand the clone dynamics of blood production, and how they might be altered in human health and diseases. Ultimately, we will be able to apply those lessons to patient care.”
Why Apex Legends’ Solo Mode Is Never Coming Back
Remember Apex Solo? It’s okay if you don’t; it might’ve been before your time. The limited-time mode (LTM) dropped alongside Season 2’s Iron Crown Collection Event. The rules are self-explanatory: drop into a Battle Royale match all by your lonesome, with 60 legends total competing for bragging rights as well as some cool character- or win-specific badges.
In some ways, playing alone was a great way to test one’s might in duels without relying on teammate assistance. However, the experimental LTM was a double-edged sword, revealing that some ability kits were simply stronger than others in 1v1s. For instance, Lifeline’s “D.O.C. Heal Drone” made her a near-unkillable force, but her “Combat Revive” passive was rendered useless since there were no allies to resurrect. You can already see some of the inconsistencies in the mode’s mechanics, right? Then it should come as no surprise that the team doesn’t plan to ever bring Apex Solo back.
Watch The Trailer That Revealed Solo Mode:
[embedded content]
While listing the myriad philosophies of character creation, lead legend designer Devan McGuire explained that solo play went against Apex’s DNA:
“This is a squad-based game, and that’s why you don’t see solos. We had that experiment a long time ago, and we’re not bringing it back. A single character should never be the answer to every problem. They should be part of that team dynamic. That’s what creates interesting strategies in the game. And we hold that as a game pillar when designing a legend: To make sure that we never break what’s core to the game. Our design pillars, the ones that we use for developing abilities and the core fundamentals of legend play patterns, are that the character needs to be ownable. It needs to occupy its own playspace amidst all the others on the roster so that it’s not stomping on the toes of someone else’s fantasy.”
Moreover, game director Steven Ferreira and design director Evan Nikolich spoke briefly about another popular LTM: Arenas. Released in Season 9 and later removed in Season 15, Arenas pitted two teams of three against one another in condensed locales. Borrowing mechanics from games like Valorant, players could buy weapons, abilities, and items from a shop at the start of each round. Arenas was so beloved that it got its own Ranked playlist; the mode’s absence is felt by many. Respawn knows this and has plans to iterate on it… someday. So, fingers crossed, we’ll see it back in the mode lineup soon.

Game Director Steven Ferreira and Design Director Evan Nikolich, Captured by Alex Van Aken
“Arenas has been backburnered,” Nikolich informed me. “But it’s in our quiver to use again at some point. When we introduced Arenas as a standalone mode, I don’t think we fully understood what we were doing in terms of bifurcating our audience. We have to build something that is more unified and brings players together to play the entirety of the game. Not just like, ‘I’m an Arenas main’ or a ‘BR main.’ It’ll potentially come back in the next 12 to 24 months. It’s definitely something on our roadmap.”
Ferreira chimed in too:
“We want to take a step back and look at what we would change about Arenas so that we won’t run into those same problems. Last year, we brought out Mixtape, and that was effectively the next evolution of Arenas. That experience was a way for players to jump in, get a lot of faster reps on weapons/legends, and just get into the core mechanics of Apex; just get a lot more cycles before they jump into the BR. As far as seeing more things outside of the BR, that’s an area that Apex is moving into. How can we continue to build on that but learn from Arenas?”
So, there you have it: Sure, no more Apex Solo, but we also haven’t seen the end of Arenas. Only time will tell when the latter LTM makes its triumphant return and how different it’ll ultimately be. If you’re not an Apex lifer just yet, check out why I think it’s still the best battle royale available right now. And trust me, it’s not even close.
Palworld Had The Biggest Third-Party Xbox Game Pass Launch Ever

Microsoft has revealed that Palworld, the “Pokémon With Guns” survival game from indie developer Pocketpair, had the biggest third-party launch ever on Xbox Game Pass. Thanks to Game Pass, Palworld has been played by more than 7 million players on Xbox, which, when added to the 12 million that have played the game on Steam, puts the total player count at 19 million. That’s a massive feat for any game, let alone an independently developed one.
🎉Total number of players exceeds 19 million🎉
It’s been less than two weeks since #Palworld was released, thank you!
・Steam: 12 million~ copies
・Xbox: 7 million~ playersWe will continue to prioritize fixing bugs!
Thank you for your continued support of #Pocketpair! pic.twitter.com/twgAeYVL07— Palworld (@Palworld_EN) January 31, 2024
Xbox shared these details about Palworld’s Game Pass launch in a new Xbox Wire post, which is also where it revealed Palworld is also the most-played third-party day-one release for Xbox Cloud Gaming (accessible with a Game Pass Ultimate subscription). The peak concurrent player count on Xbox was 3 million, “making it the most-played game on our platforms at that time,” the Xbox Wire post reads. Palworld is available for purchase in Early Access via Steam, and is available on Game Pass for both PC and Xbox in its early access equivalent Game Preview program.
“The response from fans has been tremendous and it’s incredible to see the millions of players around the world enjoying Palworld,” Pocketpair CEO Takuro Mizobe told Xbox Wire. “This is just the beginning for us and Palworld, and the feedback we’re gathering in Game Preview will allow us to continue to improve the experience for Pal Tamers across all platforms.”
[embedded content]
Pocketpair says Palworld will remain available in the Xbox Game Preview program until the team is ready for the full 1.0 release. Xbox says it’s providing support for Xbox versions of the game, including dedicated servers, engineering resource help with GPU and memory optimization, Palworld update efficiency, and general optimization.
Palworld launched earlier this month and quickly took the internet by storm. You can read about its meteoric rise and the conversation surrounding Palworld here.
For more, read the statement The Pokémon Company issued in response to Palworld’s popularity, and then check out Pocketpair’s Palworld early access roadmap. After that, read about how fake copycat versions of Palworld have started popping up on mobile app stores.
Are you playing Palworld on Xbox? Let us know how it plays in the comments below!
Imaging method reveals new cells and structures in human brain tissue

Using a novel microscopy technique, MIT and Brigham and Women’s Hospital/Harvard Medical School researchers have imaged human brain tissue in greater detail than ever before, revealing cells and structures that were not previously visible.
Among their findings, the researchers discovered that some “low-grade” brain tumors contain more putative aggressive tumor cells than expected, suggesting that some of these tumors may be more aggressive than previously thought.
The researchers hope that this technique could eventually be deployed to diagnose tumors, generate more accurate prognoses, and help doctors choose treatments.
“We’re starting to see how important the interactions of neurons and synapses with the surrounding brain are to the growth and progression of tumors. A lot of those things we really couldn’t see with conventional tools, but now we have a tool to look at those tissues at the nanoscale and try to understand these interactions,” says Pablo Valdes, a former MIT postdoc who is now an assistant professor of neuroscience at the University of Texas Medical Branch and the lead author of the study.
Edward Boyden, the Y. Eva Tan Professor in Neurotechnology at MIT; a professor of biological engineering, media arts and sciences, and brain and cognitive sciences; a Howard Hughes Medical Institute investigator; and a member of MIT’s McGovern Institute for Brain Research and Koch Institute for Integrative Cancer Research; and E. Antonio Chiocca, a professor of neurosurgery at Harvard Medical School and chair of neurosurgery at Brigham and Women’s Hospital, are the senior authors of the study, which appears today in Science Translational Medicine.
Making molecules visible
The new imaging method is based on expansion microscopy, a technique developed in Boyden’s lab in 2015 based on a simple premise: Instead of using powerful, expensive microscopes to obtain high-resolution images, the researchers devised a way to expand the tissue itself, allowing it to be imaged at very high resolution with a regular light microscope.
The technique works by embedding the tissue into a polymer that swells when water is added, and then softening up and breaking apart the proteins that normally hold tissue together. Then, adding water swells the polymer, pulling all the proteins apart from each other. This tissue enlargement allows researchers to obtain images with a resolution of around 70 nanometers, which was previously possible only with very specialized and expensive microscopes such as scanning electron microscopes.
In 2017, the Boyden lab developed a way to expand preserved human tissue specimens, but the chemical reagents that they used also destroyed the proteins that the researchers were interested in labeling. By labeling the proteins with fluorescent antibodies before expansion, the proteins’ location and identity could be visualized after the expansion process was complete. However, the antibodies typically used for this kind of labeling can’t easily squeeze through densely packed tissue before it’s expanded.
So, for this study, the authors devised a different tissue-softening protocol that breaks up the tissue but preserves proteins in the sample. After the tissue is expanded, proteins can be labelled with commercially available fluorescent antibodies. The researchers then can perform several rounds of imaging, with three or four different proteins labeled in each round. This labelling of proteins enables many more structures to be imaged, because once the tissue is expanded, antibodies can squeeze through and label proteins they couldn’t previously reach.
“We open up the space between the proteins so that we can get antibodies into crowded spaces that we couldn’t otherwise,” Valdes says. “We saw that we could expand the tissue, we could decrowd the proteins, and we could image many, many proteins in the same tissue by doing multiple rounds of staining.”
Working with MIT Assistant Professor Deblina Sarkar, the researchers demonstrated a form of this “decrowding” in 2022 using mouse tissue.
The new study resulted in a decrowding technique for use with human brain tissue samples that are used in clinical settings for pathological diagnosis and to guide treatment decisions. These samples can be more difficult to work with because they are usually embedded in paraffin and treated with other chemicals that need to be broken down before the tissue can be expanded.
In this study, the researchers labeled up to 16 different molecules per tissue sample. The molecules they targeted include markers for a variety of structures, including axons and synapses, as well as markers that identify cell types such as astrocytes and cells that form blood vessels. They also labeled molecules linked to tumor aggressiveness and neurodegeneration.
Using this approach, the researchers analyzed healthy brain tissue, along with samples from patients with two types of glioma — high-grade glioblastoma, which is the most aggressive primary brain tumor, with a poor prognosis, and low-grade gliomas, which are considered less aggressive.
“We wanted to look at brain tumors so that we can understand them better at the nanoscale level, and by doing that, to be able to develop better treatments and diagnoses in the future. At this point, it was more developing a tool to be able to understand them better, because currently in neuro-oncology, people haven’t done much in terms of super-resolution imaging,” Valdes says.
A diagnostic tool
To identify aggressive tumor cells in gliomas they studied, the researchers labeled vimentin, a protein that is found in highly aggressive glioblastomas. To their surprise, they found many more vimentin-expressing tumor cells in low-grade gliomas than had been seen using any other method.
“This tells us something about the biology of these tumors, specifically, how some of them probably have a more aggressive nature than you would suspect by doing standard staining techniques,” Valdes says.
When glioma patients undergo surgery, tumor samples are preserved and analyzed using immunohistochemistry staining, which can reveal certain markers of aggressiveness, including some of the markers analyzed in this study.
“These are incurable brain cancers, and this type of discovery will allow us to figure out which cancer molecules to target so we can design better treatments. It also proves the profound impact of having clinicians like us at the Brigham and Women’s interacting with basic scientists such as Ed Boyden at MIT to discover new technologies that can improve patient lives,” Chiocca says.
The researchers hope their expansion microscopy technique could allow doctors to learn much more about patients’ tumors, helping them to determine how aggressive the tumor is and guiding treatment choices. Valdes now plans to do a larger study of tumor types to try to establish diagnostic guidelines based on the tumor traits that can be revealed using this technique.
“Our hope is that this is going to be a diagnostic tool to pick up marker cells, interactions, and so on, that we couldn’t before,” he says. “It’s a practical tool that will help the clinical world of neuro-oncology and neuropathology look at neurological diseases at the nanoscale like never before, because fundamentally it’s a very simple tool to use.”
Boyden’s lab also plans to use this technique to study other aspects of brain function, in healthy and diseased tissue.
“Being able to do nanoimaging is important because biology is about nanoscale things — genes, gene products, biomolecules — and they interact over nanoscale distances,” Boyden says. “We can study all sorts of nanoscale interactions, including synaptic changes, immune interactions, and changes that occur during cancer and aging.”
The research was funded by K. Lisa Yang, the Howard Hughes Medical Institute, John Doerr, Open Philanthropy, the Bill and Melinda Gates Foundation, the Koch Institute Frontier Research Program, the National Institutes of Health, and the Neurosurgery Research and Education Foundation.
